Category:Media from Justine et al. 2022 - PeerJ/12725
Media from:
Jean-Lou Justine, Romain Gastineau, Pierre Gros, Delphine Gey, Enrico Ruzzier, Laurent Charles, & Leigh Winsor
(2022-02-01). "Hammerhead flatworms (Platyhelminthes, Geoplanidae, Bipaliinae): mitochondrial genomes and description of two new species from France, Italy, and Mayotte". PeerJ 10: e12725. PeerJ. DOI:10.7717/peerj.12725. ISSN 2167-8359.
scientific article | |||||
| Upload media | |||||
| Instance of | |||||
|---|---|---|---|---|---|
| Main subject | |||||
| Author |
| ||||
| Published in | |||||
| Full work available at URL | |||||
| Copyright license | |||||
| Publication date |
| ||||
| |||||
Abstract
Background
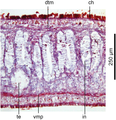
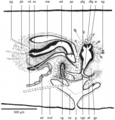
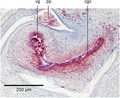
New records of alien land planarians are regularly reported worldwide, and some correspond to undescribed species of unknown geographic origin. The description of new species of land planarians (Geoplanidae) should classically be based on both external morphology and histology of anatomical structures, especially the copulatory organs, ideally with the addition of molecular data.
Methods Here, we describe the morphology and reproductive anatomy of a species previously reported as Diversibipalium “black”, and the morphology of a species previously reported as Diversibipalium “blue”. Based on next generation sequencing, we obtained the complete mitogenome of five species of Bipaliinae, including these two species.
Results The new species Humbertium covidum n. sp. (syn: Diversibipalium “black” of Justine et al., 2018) is formally described on the basis of morphology, histology and mitogenome, and is assigned to Humbertium on the basis of its reproductive anatomy. The type-locality is Casier, Italy, and other localities are in the Department of Pyrénées-Atlantiques, France; some published or unpublished records suggest that this species might also be present in Russia, China, and Japan. The mitogenomic polymorphism of two geographically distinct specimens (Italy vs France) is described; the cox1 gene displayed 2.25% difference. The new species Diversibipalium mayottensis n. sp. (syn: Diversibipalium “blue” of Justine et al., 2018) is formally described on the basis of external morphology and complete mitogenome and is assigned to Diversibipalium on the basis of an absence of information on its reproductive anatomy. The type- and only known locality is the island of Mayotte in the Mozambique Channel off Africa. Phylogenies of bipaliine geoplanids were constructed on the basis of SSU, LSU, mitochondrial proteins and concatenated sequences of cox1, SSU and LSU. In all four phylogenies, D. mayottensis was the sister-group to all the other bipaliines. With the exception of D. multilineatum which could not be circularised, the complete mitogenomes of B. kewense, B. vagum, B. adventitium, H. covidum and D. mayottensis were colinear. The 16S gene in all bipaliine species was problematic because usual tools were unable to locate its exact position.
Conclusion Next generation sequencing, which can provide complete mitochondrial genomes as well as traditionally used genes such as SSU, LSU and cox1, is a powerful tool for delineating and describing species of Bipaliinae when the reproductive structure cannot be studied, which is sometimes the case of asexually reproducing invasive species. The unexpected position of the new species D. mayottensis as sister-group to all other Bipaliinae in all phylogenetic analyses suggests that the species could belong to a new genus, yet to be described.
Media in category "Media from Justine et al. 2022 - PeerJ/12725"
The following 41 files are in this category, out of 41 total.
-
Fig-01-full Humbertium covidum Justine et al PeerJ 2022.png 2,430 × 1,230; 94 KB
-
Fig-02-full Humbertium covidum Justine et al PeerJ 2022.png 1,868 × 1,051; 4.11 MB
-
Fig-03-full Humbertium covidum Justine et al PeerJ 2022.png 1,868 × 1,051; 3.3 MB
-
Fig-04-full Humbertium covidum Justine et al PeerJ 2022.png 1,869 × 1,051; 3.56 MB
-
Fig-05-full Humbertium covidum Justine et al PeerJ 2022.png 1,868 × 1,051; 3.24 MB
-
Fig-06-full Humbertium covidum Justine et al PeerJ 2022.png 1,868 × 1,051; 4.02 MB
-
Fig-07-full Humbertium covidum Justine et al PeerJ 2022.png 1,868 × 1,051; 2.72 MB
-
Fig-08-full Humbertium covidum Justine et al PeerJ 2022.png 1,868 × 1,051; 2.64 MB
-
Fig-09-full Humbertium covidum Justine et al PeerJ 2022.png 1,868 × 1,868; 4.81 MB
-
Fig-10-full Humbertium covidum Justine et al PeerJ 2022.png 1,883 × 1,066; 3.18 MB
-
Fig-11-full Humbertium covidum Justine et al PeerJ 2022.png 1,868 × 1,868; 2.81 MB
-
Fig-12-full Humbertium covidum Justine et al PeerJ 2022.png 1,868 × 1,862; 4.05 MB
-
Fig-13-full Humbertium covidum (anatomy) Justine et al PeerJ 2022.png 1,883 × 1,247; 3.47 MB
-
Fig-14-full Humbertium covidum (anatomy) Justine et al PeerJ 2022.png 1,883 × 1,946; 4.75 MB
-
Fig-15-full Humbertium covidum (anatomy) Justine et al PeerJ 2022.png 1,883 × 2,167; 4.59 MB
-
Fig-16-full Humbertium covidum (anatomy) Justine et al PeerJ 2022.png 1,509 × 1,584; 60 KB
-
Fig-17-full Humbertium covidum (anatomy) Justine et al PeerJ 2022.png 1,883 × 1,957; 1.62 MB
-
Fig-18-full Humbertium covidum (anatomy) Justine et al PeerJ 2022.png 1,883 × 1,595; 5.42 MB
-
Fig-19-full Humbertium covidum (anatomy) Justine et al PeerJ 2022.png 1,883 × 1,975; 6.1 MB
-
Fig-20-full Humbertium covidum (anatomy) Justine et al PeerJ 2022.png 1,883 × 1,884; 5.96 MB
-
Fig-21-full Humbertium covidum (anatomy) Justine et al PeerJ 2022.png 1,883 × 1,560; 5.08 MB
-
Fig-22-full Humbertium covidum (anatomy) Justine et al PeerJ 2022.png 1,884 × 1,541; 5.13 MB
-
Fig-23-full Humbertium covidum (anatomy) Justine et al PeerJ 2022.png 1,883 × 1,695; 4.26 MB
-
Fig-24-full Diversibipalium mayottensis Justine et al PeerJ 2022.png 1,868 × 1,870; 6.62 MB
-
Fig-25-full Diversibipalium mayottensis Justine et al PeerJ 2022.png 1,883 × 1,885; 6.32 MB
-
Fig-26-full Diversibipalium mayottensis Justine et al PeerJ 2022.png 1,868 × 1,864; 5.69 MB
-
Fig-27-full Diversibipalium mayottensis Justine et al PeerJ 2022.png 1,868 × 1,876; 5.86 MB
-
Fig-28-full Mitogenome Humbertium covidum Justine et al PeerJ 2022.png 3,355 × 3,357; 2.74 MB
-
Fig-29-full Mitogenome Humbertium covidum Justine et al PeerJ 2022.png 3,355 × 3,355; 2.67 MB
-
Fig-30-full Mitogenome Diversibipalium mayottensis Justine et al PeerJ 2022.png 3,355 × 3,353; 2.15 MB
-
Fig-31-full Mitogenome Bipalium vagum Justine et al PeerJ 2022.png 3,355 × 3,353; 2.7 MB
-
Fig-32-full Mitogenome Bipalium adventitium Justine et al PeerJ 2022.png 3,355 × 3,338; 2.97 MB
-
Fig-33-full Mitogenome Diversibipalium multilineatum Justine et al PeerJ 2022.png 3,354 × 1,176; 228 KB
-
Fig-34-full Mitogenome Bipalium kewense Justine et al PeerJ 2022.png 3,355 × 3,355; 2.09 MB
-
Fig-35-full LOGO Alignment Justine et al PeerJ 2022.png 3,355 × 2,555; 8.61 MB
-
Fig-36-full 16S Alignment Justine et al PeerJ 2022.png 3,356 × 1,565; 1.46 MB
-
Fig-37-full SSU Tree Justine et al PeerJ 2022.png 2,430 × 1,886; 155 KB
-
Fig-38-full LSU Tree Justine et al PeerJ 2022.png 2,430 × 1,635; 210 KB
-
Fig-39-full Mitochondrial proteins tree Justine et al PeerJ 2022.png 2,420 × 1,531; 281 KB
-
Fig-40-full Three-gene tree Justine et al PeerJ 2022.png 2,436 × 1,190; 189 KB
-
Humbertium covidum with scale at end mm.webm 1 min 4 s, 960 × 540; 10.48 MB