File:Atlanta serial killer 1979 1981 cost analysis barycenter points weighted avg rasters 1 1.png

Original file (992 × 744 pixels, file size: 571 KB, MIME type: image/png)
Captions
Captions
Summary
edit| DescriptionAtlanta serial killer 1979 1981 cost analysis barycenter points weighted avg rasters 1 1.png |
English: Cost analysis of Atlanta serial killer 1979-1981: barycenter and sites weaighted average |
| Date | |
| Source | Own work |
| Author | Merikanto |
Stamen map w/leaflet r
Uses Stamen Toner no symbols map
https://github.com/stamen/maps.stamen.com/blob/master/LICENSE
Copyright (c) 2012, Stamen Design
All rights reserved.
Redistribution and use in source and binary forms, with or without
modification, are permitted provided that the following conditions are met:
* Redistributions of source code must retain the above copyright
notice, this list of conditions and the following disclaimer.
* Redistributions in binary form must reproduce the above copyright
notice, this list of conditions and the following disclaimer in the
documentation and/or other materials provided with the distribution.
* Neither the name of Stamen Design nor the names of its contributors may
be used to endorse or promote products derived from this software without
specific prior written permission.
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND
ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED
WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
DISCLAIMED. IN NO EVENT SHALL <COPYRIGHT HOLDER> BE LIABLE FOR ANY
DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES
(INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES;
LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND
ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT
(INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS
SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
R code to download stamen toner no symbols , main roads etc, and make distance raster and tweaked accumulated cost map from them
-
- cost analysis raster
- barycenter cost and all points costs rasters wwaighted avg
- user data:
- input rasters distance from main roads
- input locations
-
- 11.0.2023 0000.0001
-
- based on
-
- load map tiles, process them with magick
-
- make black white thinned road map from stamen toner
-
library(tidyverse)
library(sf)
library(sp)
library(raster)
library(rgdal)
library(rgeos)
library(fasterRaster)
library(rgrass)
library(magick)
library(terra)
library(gdistance)
library(maptiles)
- map view
library(leaflet)
library(pandoc)
library(htmlwidgets)
library(webshot)
library(mapview)
load_tiles_from_net=0 ## load stamen toner no symbols
process_raster=0 ## make distance from main roads (and other stuff)
produce_map=1 ## create cost raster and map
dimx=256
dimy=256
- boxi1 <- st_read("./gisa1/atlanta1.shp")
clat1 <- c(33.754065)
clon1 <- c(-84.446577)
delta1=0.4
crimelats<-c(33.703093,
33.660032,
33.741141,
33.711061,
33.701493,
33.746652,
33.6605585,
33.755227,
33.692281,
33.738875,
33.805397,
33.677783,
33.679033,
33.858629,
33.68205,
33.68164,
33.837342,
33.680747,
33.631259,
33.683782,
33.653495,
33.766465,
33.653852,
33.802901,
33.7392226,
33.804113)
crimelons<- c(
-84.532406,
-84.49509,
-84.383959,
-84.447227,
-84.584169,
-84.350482,
-84.4941276,
-84.465294,
-84.350066,
-84.408613,
-84.470401,
-84.427292,
-84.358048,
-84.455166,
-84.573247,
-84.067554,
-84.332364,
-84.249387,
-84.128966,
-84.64159,
-84.681008,
-84.422132,
-84.6803,
-84.500141,
-84.3287373,
-84.499154)
- crimelocations = np.array([[33.703093, -84.532406], [33.660032, -84.49509], [33.741141, -84.383959], [33.711061, -84.447227], [33.701493, -84.584169], [33.746652, -84.350482], [33.6605585, -84.4941276], [33.755227, -84.465294], [33.692281, -84.350066], [33.738875, -84.408613], [33.805397, -84.470401], [33.677783, -84.427292], [33.679033, -84.358048], [33.858629, -84.455166], [33.68205, -84.573247], [33.68164, -84.067554], [33.837342, -84.332364], [33.680747, -84.249387], [33.631259, -84.128966], [33.683782, -84.64159], [33.653495, -84.681008], [33.766465, -84.422132], [33.653852, -84.6803], [33.802901, -84.500141], [33.7392226, -84.3287373], [33.804113, -84.499154]])
suspectlat <- c(33.754065)
suspectlon <- c(-84.446577)
p1=c(-84.532406,33.703093)
p2=c(-84.49509,33.660032)
p3=c(-84.383959,33.741141)
p4= c(-84.447227,33.711061)
p5= c(-84.584169,33.701493)
p6= c(-84.350482,33.746652)
p7= c(-84.4941276,33.6605585)
p8= c(-84.465294,33.755227)
p9= c(-84.350066,33.692281)
p10= c(-84.408613,33.738875)
p11= c(-84.470401,33.805397)
p12= c(-84.427292,33.677783)
p13= c(-84.358048,33.679033)
p14= c(-84.455166,33.858629)
p15= c(-84.573247,33.68205)
p16= c(-84.067554,33.68164)
p17= c(-84.332364,33.837342)
p18= c(-84.249387,33.680747)
p19= c(-84.128966,33.631259)
p20= c(-84.64159,33.683782)
p21= c(-84.681008,33.653495)
p22= c(-84.422132,33.766465)
p23= c(-84.6803,33.653852)
p24= c(-84.500141,33.802901)
p25= c(-84.3287373,33.7392226)
p26= c(-84.499154,33.804113)
minlat1=min(clat1)+delta1
maxlat1=max(clat1)-delta1
minlon1=min(clon1)+delta1
maxlon1=max(clon1)-delta1
nn1=maxlat1
ss1=minlat1
ww1=minlon1
ee1=maxlon1
-
- https://rdrr.io/github/adamlilith/fasterRaster/man/fasterVectorize.html
-
- you must manually set grass 7.8 path
- note easier not use osgeow grass, use standalone grass 7.8
- grassDir <-'C:/Program Files/GRASS GIS 7.8'
grassDir <-'C:/Program Files/GRASS GIS 8.3'
- execGRASS("g.list", parameters = list(type = "vector"))
- execGRASS("g.distance")
- stop(-1)
boxi0 <- rgeos::bbox2SP(n = nn1, s = ss1, w = ww1, e = ee1,
proj4string = CRS("+proj=longlat +ellps=WGS84 +datum=WGS84 +no_defs"))
boxi1=st_bbox(boxi0)
- plot(boxi1)
- stop(-1)
if(load_tiles_from_net==1) {
print("Trying to load map tiles ...")
maptiles1 <- get_tiles(x = boxi1, provider = "Stamen.TonerBackground", crop = TRUE,
zoom = 11, verbose = TRUE)
# Plot the tiles
#plot_tiles(maptiles1)
#str(maptiles1)
rout1<-as(maptiles1, "Raster")
#plot(rout1)
#extent(r2)<-extent1
crs(rout1) <- "+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0"
f <- writeRaster(rout1, "atlas1.tif", format="Gtiff", overwrite=TRUE)
#plot(rout1)
#image(rout1)
print("Tiles loaded.")
}
if(process_raster==1)
{
rin1<-raster("atlas1.tif")
sab1<-raster(nrow=dimx,ncol=dimy)
extent(sab1)<-extent(rin1)
crs(sab1)<-crs(rin1)
rin2<-resample(rin1, sab1)
rin2<-255-rin2
rin2[rin2>127]=127
rin2[rin2>4]=4
rin2[rin2<4]=0
rin2[rin2>0]=1
rin2[rin2<1]=0
rin3<-rin2
print("*")
- plot(rin3)
- rin3<-rin3-1
rout1<-rin3
crs(rout1) <- "+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0"
plot(rout1)
f <- writeRaster(rout1, "muva1.tif", format="Gtiff", overwrite=TRUE)
- f <- writeRaster(rout1, "muva1.png", format="png", overwrite=TRUE)
- tiger <- image_read_svg('http://jeroen.github.io/images/tiger.svg', width = 350)
- print(tiger)
- image_write(tiger, path = "tiger.png", format = "png")
inras1<-raster("muva1.tif")
- str(inras1)
- stop(-1)
ext1<-extent(inras1)
- crs1<-srs(inras1)
in1 <- image_read('muva1.tif')
- plot(in1)
- in1<-image_negate(in1)
- ima1<-in1
- in1<-in1-1
plot(in1)
ima1<-in1 %>% image_morphology('Thinning', 'Skeleton', iterations = 5)
- ima1 <- in1 %>% image_morphology('Dilate', "Plus", iterations = 1)
plot(ima1)
- stop(-1)
- str(ima1)
- rout1<-raster::raster(ima1)
rout0 <- as.raster(ima1) |> as.matrix() |> rast()
rout1<-brick(rout0)
crs1 ="+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0"
str(rout1)
crs(rout1)<-crs1
extent(rout1)<-ext1
rout1<-rout1-1
plot(rout1)
f <- writeRaster(rout1, "thinned1.tif", format="Gtiff", overwrite=TRUE)
rin4<-raster("thinned1.tif")
- print(" Vectorize ...")
- stop(-1)
- could also use rasterToPolygons() which is
- probably faster in this example
- forestPoly <- fasterVectorize(rast=madForest2000,vectType='area', grassDir=grassDir)
- polys1<- rasterToPolygons(rin4, dissolve=TRUE)
- par(mfrow=c(1, 2))
- sf1 = st_as_sf(lines11, coords = c("x", "y"), crs = 4326, agr = "constant")
- st_write(sf1, "vektor.geojson")
- str(rout1)
rin1<-raster("./thinned1.tif")
sabluna<- raster(nrow=dimx, ncol=dimy)
ext1<-extent(rin1)
crs1<-crs(rin1)
extent(sabluna)<-ext1
crs(sabluna)<-crs1
r <- resample(rin1,sabluna, method='bilinear')
r[r>0]=1
r[r<1]=0
- plot(r)
- stop(-1)
- r[r==1]=NA
- plot(r)
- dis1 <- raster::distance(r, doEdge = T)
- calculate distance
dis1 <- gridDistance(r, 1, omit=-1)
plot(dis1)
rout1<-dis1
extent(rout1)<-ext1
crs(rout1) <- "+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0"
f <- writeRaster(rout1, "./distance1.tif", format="GTiff", overwrite=TRUE)
} ## process stmen toner map raster
if(produce_map==1)
{
- distance raster input
- made with qgis
rin1<-raster("./gisa1/distance1.tif")
sabluna<- raster(nrow=dimx, ncol=dimy)
ext1<-extent(rin1)
crs1<-crs(rin1)
extent(sabluna)<-ext1
crs(sabluna)<-crs1
r0 <- resample(rin1,sabluna, method='bilinear')
max1=maxValue(r0)
min1=minValue(r0)
delta1=max1-min1
r<-(r0-min1)/delta1
plot(r)
- stop(-1)
centerlat1=mean(crimelats)
centerlon1=mean(crimelons)
- print(centerlat1, centerlon1)
- p1 <- SpatialPoints( rbind( c(-84.532406, 33.703093) ))
- p1<- SpatialPoints( rbind( c(crimelons, crimelats) ))
pon2<- rbind(p1,p2,p3,p4,p5,p6,p7,p8,p9,p10,p11,p12,p13,p14,p15,p16,p17,p18,p19,p20,p21,p22,p23,p24,p25,p26)
p1<-c(centerlon1, centerlat1)
pon3<-p1
- coords1=c()
T1<- transition(r, function(x) 1/mean(x), 8)
- T1 <- transition(r, function(x) 1/mean(x), 8)
- 1/mean: reciprocal to get permeability
T1 <- geoCorrection(T1)
r2 <- accCost(T1, pon2)
r3 <- accCost(T1, pon3)
plot(r2)
plot(r3)
- stop(-1)
r2=r2/100 ## many points
r3=r3/100 ## barycenter
r2b=r2
- r2b=exp(-r2b)
r3b=exp(-r3*0.66)
max1=maxValue(r3b)
min1=minValue(r3b)
delta1=max1-min1
r3b<-(r3b-min1)/delta1
- r2b[r2b >1] <-exp(-r3)
r2b=(sin(r2b*1)+1)/2
r2c=exp(-r2*0.66)
max1=maxValue(r2c)
min1=minValue(r2c)
delta1=max1-min1
r2c<-(r2c-min1)/delta1
r2d=r2b*r2c
r4<-(r2d*3+r3b)/4
- r4<-r2b
plot(r4)
- text(A1)
extent(r4)<-ext1
crs(r4) <- "+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0"
r5<-r4*100
f <- writeRaster(r5, "accostmap1.tif", format="Gtiff", overwrite=TRUE)
delta1=0.05
minlat1=min(crimelats)+delta1
maxlat1=max(crimelats)-delta1
minlon1=min(crimelons)+delta1
maxlon1=max(crimelons)-delta1
extent1<-c(minlon1, maxlon1, minlat1, maxlat1)
- Stadia.StamenTonerBackground
- tiles1="https://server.arcgisonline.com/ArcGIS/rest/services/World_Imagery/MapServer/tile/{z}/{y}/{x}.png" ## satellite
- tiles1="https://{s}.tile-cyclosm.openstreetmap.fr/cyclosm/{z}/{x}/{y}.png"
- tiles1="https://server.arcgisonline.com/ArcGIS/rest/services/World_Street_Map/MapServer/tile/{z}/{y}/{x}.png"
- tiles1="https://server.arcgisonline.com/ArcGIS/rest/services/Canvas/World_Light_Gray_Base/MapServer/tile/{z}/{y}/{x}.png"
- tiles1="https://{s}.basemaps.cartocdn.com/light_all/{z}/{x}/{y}{r}.png"
- tiles1="https://{s}.basemaps.cartocdn.com/light_nolabels/{z}/{x}/{y}{r}.png"
- tiles1="https://{s}.basemaps.cartocdn.com/dark_nolabels/{z}/{x}/{y}{r}.png"
- tiles1=https://{s}.basemaps.cartocdn.com/rastertiles/voyager_nolabels/{z}/{x}/{y}{r}.png"
tiles1="https://tiles.stadiamaps.com/tiles/stamen_toner_background/{z}/{x}/{y}{r}.png" #
contours1 <- rasterToContour(r5)
r6<-r5*1
pal = colorNumeric(c("blue", "green", "yellow", "orange", "red", "violet", "violet"), 50:100,na.color = "transparent")
map1 <- leaflet(padding=0) %>%
addTiles(tiles1) %>%
fitBounds(minlon1,minlat1,maxlon1,maxlat1) %>%
setView(-84.39, 33.755, zoom = 11) %>%
addRasterImage(r6, colors = pal, opacity = 0.5) %>%
#addRasterImage(r6, opacity = 0.5) %>%
addPolylines(data =contours1,fillOpacity=0.5,fillColor = "transparent",opacity=0.5,weight=1) %>%
addCircleMarkers(lng = crimelons, lat = crimelats, radius = 8, color = "#F00", popup = NULL) %>%
fitBounds(minlon1,minlat1,maxlon1,maxlat1) %>%
addCircleMarkers(suspectlon, suspectlat, radius = 10, color = "#003", popup = NULL)
- addMarkers(suspectlon, suspectlat)
- addMarkers(crimelons, crimelats)
- map1<-fitBounds(minlon1,minlat1,maxlon1,maxlat1)
saveWidget(map1, file="map1.html", selfcontained = FALSE)
- webshot("map1.html", file = "map1.png",cliprect = "viewport", remove_controls = c("zoomControl", "layersControl", "homeButton", "scaleBar",
- "drawToolbar", "easyButton") )
- mapshot(map1, file = paste0(getwd(), "/map1.png"))
willremove1<-c("zoomControl", "layersControl", "homeButton", "scaleBar",
"drawToolbar", "easyButton")
mapshot(map1, file = paste0(getwd(), "./map1.png"),remove_controls = willremove1)
r1<-raster("./map1.png")
r2<-r1
extent(r2)<-extent1
crs(r2) <- "+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0"
f <- writeRaster(r2, "map1.tif", format="Gtiff", overwrite=TRUE)
print(".")
}
Licensing
edit- You are free:
- to share – to copy, distribute and transmit the work
- to remix – to adapt the work
- Under the following conditions:
- attribution – You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
- share alike – If you remix, transform, or build upon the material, you must distribute your contributions under the same or compatible license as the original.
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 09:02, 11 September 2023 |  | 992 × 744 (571 KB) | Merikanto (talk | contribs) | Update |
| 14:31, 10 September 2023 | 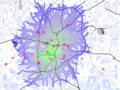 | 992 × 744 (637 KB) | Merikanto (talk | contribs) | Uploaded own work with UploadWizard |
You cannot overwrite this file.
File usage on Commons
There are no pages that use this file.
Metadata
This file contains additional information such as Exif metadata which may have been added by the digital camera, scanner, or software program used to create or digitize it. If the file has been modified from its original state, some details such as the timestamp may not fully reflect those of the original file. The timestamp is only as accurate as the clock in the camera, and it may be completely wrong.
| Horizontal resolution | 37.8 dpc |
|---|---|
| Vertical resolution | 37.8 dpc |